A contextual genomic perspective on physical activity and its relationship to health, well being and illness
MVP datasets
The U.S. Department of Veterans Affairs (VA) MVP is one of the largest and most diverse biobanks in the world, with genetic and electronic health record (EHR) data available45,46. Ethical approval for the MVP study was obtained from the Central VA Institutional Review Board and the site-specific institutional review boards. All relevant ethical regulations for work with human participants were followed in the conduct of the study, and informed consent was obtained from all participants.
Participants provided a blood sample for genomic analyses, granted access to medical records, and a subset agreed to complete two questionnaires, the MVP Baseline and Lifestyle Surveys. PA information used in this study was collected from the MVP Lifestyle Survey, and data were subdivided according to context (leisure, work and home), intensity (vigorous, moderate and light) and frequency (Supplementary Fig. 1). The three levels of activity were defined as follows:
-
1.
Vigorous—activities that cause your heart to beat rapidly and you work up a good sweat, and are breathing heavily; performed at least 10 min at a time.
-
2.
Moderate—activities that cause your heart rate to increase slightly and you typically work up a sweat, but are not physically exhausting; performed for at least 10 min at a time.
-
3.
Light—activities that require little physical effort.
The levels of activity were considered together with frequency information, which could be ‘daily’, ‘several times per week’, ‘once per week’, ‘several times per month’, ‘once per month or less’ or ‘never’, when participants answered three questions regarding the place where PA was effected (Supplementary Methods). The three different levels of PA were summed considering All-PA phenotype frequency and weighting intensity to create a total score, reported in the Supplementary Methods.
From previous MVP analyses, after MVP genotyping performed using a customized Affymetrix Axiom Biobank Array and quality control47, EAGLE2 (ref. 48) was used for phasing chromosomes and Minimac3 was used for imputation49, using the 1000 Genomes Project reference panel, Phase 3, version 5 (ref. 50). Populations were defined using principal component analysis45.
For All-PA during leisure time (All-PA–leisure), we ran independent GWAS for each of the three ancestries (EUR, AFR and AMR; Supplementary Fig. 1). For EUR, we also ran GWAS for vigorous PA during leisure time (Vig-PA–leisure), work time (Vig-PA–work) and home time (Vig-PA–home). We did not have enough power to run GWAS for vigorous PA for AFR and AMR ancestries.
In the quality control procedure for GWAS conducted using PLINK 2.0 (ref. 51), we removed variants with imputation quality scores < 0.6, Hardy–Weinberg equilibrium P < 5 × 10−5, minor allele frequency <0.01, missing call rates for variants >0.1 and missing call rates for samples >0.1. Data were aligned to the GRCh37 reference genome. Considering EUR ancestry, after filtering, we had 6,660,206 variants for All-PA–leisure phenotype, and 6,548,052, 6,536,056, 6,537,158 variants for Vig-PA–leisure, Vig-PA–home and Vig-PA–work, respectively. For AFR and AMR ancestries, we kept 11,984,943 and 7,969,366 variants, respectively (All-PA–leisure phenotype).
To remove related individuals, we used a threshold of 0.0884 for the kinship coefficients calculated by KING52, resulting in the removal of individuals with a minimum of a second-degree relationship. Then, we implemented an algorithm to optimize retaining the more informative individuals, keeping the maximum number of individuals with the highest score (for All-PA–leisure phenotype) or with the highest frequency (for the vigorous activity phenotypes). If two individuals had the same score (or frequency), we removed the one with the highest number of relationships. For EUR, the final sample sizes were as follows: 189,812 individuals for the All-PA–leisure phenotype, 201,050 for Vig-PA–leisure, 203,430 for Vig-PA–home and 171,278 for Vig-PA–work. For AFR and AMR ancestries, we kept 27,044 and 10,263 individuals, respectively (All-PA–leisure phenotype). We ran GWAS analysis using a linear regression model implemented in PLINK 2.0, using sex, age and the first ten principal components as covariates.
For All-PA–leisure&home phenotype and for All-PA–leisure&home&work, we ran the same QC as described in the Methods, using EUR ancestry. After filtering, we had 6,678,873 variants for All-PA–leisure&home, and 6,761,070 variants for All-PA–leisure&home&work. For the final sample size, where we excluded individuals without full information on their PA, we retained 181,317 individuals for All-PA–leisure&home, and 146,929 for All-PA–leisure&home&work.
UKB summary statistics
UKB is a cohort study of ~500,000 adults aged 40–69 years in the United Kingdom (UK) and was recruited from 22 centers across the country53. All UKB participants provided written informed consent53. Ethical approval of the UKB study was given by the North West Multicentre Research Ethics Committee, the National Information Governance Board for Health and Social Care and the Community Health Index Advisory Group53.
From the UKB public databases, we downloaded GWAS summary statistics for the SSOE phenotype with the accession code GCST006100, for MVPA with the accession code GCST006097 and for VPA with the accession code GCST006098 (ref. 13). Self-reported information was reported via a touchscreen questionnaire. For SSOE, participants needed to answer the following question: ‘in the last 4 weeks, did you spend any time doing the following?’. Then, there were follow-up questions assessing the frequency and typical duration of ‘strenuous sports’ and of ‘other exercises’. Cases were defined as individuals spending 2–3 days per week or more doing strenuous sports or other exercises for a duration of 15–30 min or greater, and controls as those individuals who did not indicate spending any time in the last 4 weeks doing either SSOE, obtaining 124,842 cases (ncase) and 225,650 controls (ncontrol)13. We defined the population effective size as neff = 4/((1/ncase) + (1/ncontrol)).
The downloaded summary statistics had SNPs filtered according to—Hardy–Weinberg equilibrium P < 10−6, high missingness >1.5%, low minor allele frequency <0.1%13. We also filtered low imputation quality <0.6, yielding approximately 11.7 million available SNPs.
Meta-analyses
METAL54 was used for EUR ancestry meta-analysis, which included EUR MVP data for the All-PA–leisure phenotype and UKB summary statistics for the SSOE phenotype13, in an All-PA–leisure + SSOE meta-analysis. We also ran a cross-ancestry All-PA–leisure + SSOE meta-analysis, adding AFR and AMR summary statistics from MVP data to the EUR All-PA–leisure + SSOE meta-analysis (Supplementary Fig. 1). We conducted the meta-analyses using the sample-size method, which considers P value and direction of effect, weighted according to sample size. We used this method to combine summary statistics from a logistic regression model (UKB summary statistics for SSOE phenotype) and a linear regression model (MVP summary statistics for All-PA–leisure phenotype).
XWAS
Sex-stratified analysis of the X chromosome was conducted in the EUR MVP sample using XWAS 3.0 on hard-call genotypes55. We included 174,375 males and 15,130 females for the All-PA–leisure phenotype and 229,331 males and 17,072 females for Vig-PA–leisure. Following sex-stratified analysis, test statistics from males and females were combined using Stouffer’s method56. Variants with Hardy–Weinberg equilibrium P < 5 × 10−6 were filtered; 191,114 variants were included for the All-PA–leisure phenotype and 191,148 for Vig-PA–leisure. We used age and the first ten principal components as covariates.
Gene-based and gene-set analyses, functional enrichment
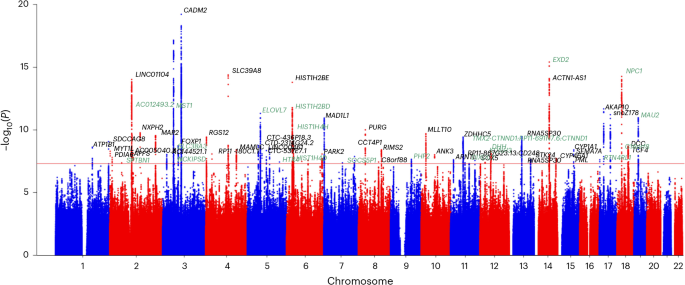
Using the tool MAGMA57, implemented in the FUMA web-based platform58, we annotated variants and ran gene-based tests, gene-set analyses and functional enrichment for each of the two meta-analyses (EUR ancestry and cross-ancestry) for the All-PA–leisure phenotype and for the GWAS of Vig-PA–leisure. For annotation, we used 1000 Genome Phase 3 EUR for EUR ancestry analyses and 1000 Genome Phase 3 ALL for the cross-ancestry All-PA–leisure+SSOE meta-analysis. The other parameters for the identification of lead SNPs were a maximum cutoff P < 0.05, r2 threshold to define independent significant SNPs >0.6, a second r2 threshold to define lead SNPs >0.1 and a maximum distance between linkage disequilibrium (LD) blocks to merge into a locus <250 kb.
The MAGMA gene-based test is based on a multiple regression approach to detect gene effects, considering SNP P values and linkage disequilibrium. We also ran a gene-based approach on our EUR All-PA–leisure + SSOE meta-analysis using fastENLOC17,18 (fast enrichment estimation aided colocalization analysis). This method was used to prioritize likely causal gene–trait associations with precomputed eQTL annotations from the 49 tissues in the GTEx v8 dataset. We kept as ‘noteworthy genes’ those with GLCP ≥ 0.9; if a gene was colocalizing among two or more tissues, we retained the tissue with the highest GLCP value.
Gene-set analyses are performed for curated gene sets and GO terms obtained from MsigDB, with terms having a Bonferroni-corrected P < 0.05 considered significant57. Gene property analyses were performed for tissue-specific gene expression using GTEx v8 dataset59.
Single-cell expression
FUMA also includes a tool to test cell-type-specificity analysis60, which was run using the following datasets of human samples: Allen Brain Atlas Cell Type61, DroNc62, GSE104276 (ref. 63), GSE67835 without fetal samples64, GSE81547 (ref. 65), GSE84133 (ref. 66), GSE89232 (ref. 67), GSE101601 (Linnarsson’s lab)68, GSE76381 (Linnarsson’s lab)69 and PsychENCODE70.
SNP-h
2 and genetic correlation
LDSC16 was used to calculate SNP-h2 for EUR ancestry and to calculate genetic correlation71 between the EUR meta-analysis of the All-PA–leisure phenotype and other traits, to quantify genetic similarity (Supplementary Methods ‘Traits studied for genetic correlation with PA’). To determine which traits were significant, we used the Benjamini–Hochberg procedure (Supplementary Table 15; ‘Benjamini–Hochberg false discovery procedure’). To estimate SNP-h2 for AFR and AMR cohorts, we calculated LD scores with cov-LDSC72 from 10,000 random independent individuals from MVP filtering the SNPs to keep only those that were previously identified in the HapMap Project73. We conducted multitrait conditional and joint analysis to evaluate the potential confounding of socioeconomic status with mtCOJO (Supplementary Notes).
Benjamini–Hochberg false discovery procedure
To determine which traits were significantly genetically correlated with PA, we used the Benjamini–Hochberg procedure (Supplementary Table 19)74. We ranked the traits according to their increasing P value, then we calculated the P value’s Benjamini–Hochberg critical value by multiplying each rank by 0.05 and dividing it by the total number of analyzed traits. Finally, the trait P values were compared to the critical values, considering a trait significant until the largest P value was smaller than its corresponding critical value.
Local genetic correlation
We ran LAVA19 to calculate local genetic correlations between the EUR All-PA–leisure + SSOE meta-analysis and the 41 traits (Supplementary Table 41) also studied for genome-wide (global) genetic correlation, to identify shared genetic bases in specific genomic regions. LAVA allows estimation of local genetic heritability and local genetic correlations. We considered 2,495 genomic loci, defined by partitioning the genome into blocks of around 1 Mb and minimizing the LD between the blocks19. Based on the number of genomic loci, we defined significant local genetic heritability as having a Bonferroni-corrected P < 0.05/2,495. We evaluated local genetic correlation at loci with pairs of traits that have already been shown to have significant local genetic heritability for both phenotypes. Thus, we conducted 3,088 local genetic correlation tests, defining significance as those with a Bonferroni-corrected P < 0.05/3,088.
MR
MR analyses enable the inference of causality between traits with genetic similarity. Two-sample MR was performed using the following four methods: MR-Egger, weighted median, IVW and simple mode75. For the PA trait, we used EUR ancestry MVP data of the All-PA–leisure phenotype to account for problems of population stratification and of overlapping samples. We filtered SNPs with P > 10−5 and clumping data using the default window of 10,000 kb and the r2 cutoff of 0.001. To designate significant results, we applied multiple testing correction for 36 outcomes used both as outcomes and as exposures (P = 6.9 × 10−4). We also used TwoSampleMR to run MVMR analyses of the All-PA–leisure phenotype as exposure, to understand the influence that BMI could have as a confounder, setting it as exposure (Supplementary Notes).
TWAS
TWAS was used to identify genes associated to traits. We ran TWAS using FUSION76 with the 1000 Genomes LD reference data ( and the GTEx v8 multitissue expression weights from 49 tissues ( These tissues included adipose, adrenal gland, artery, brain, breast, skin, blood, colon, esophagus, heart, kidney, liver, lung, minor salivary gland, muscle, nerve, ovary, pancreas, pituitary, prostate, small intestine, spleen, stomach, testis, thyroid, uterus and vagina. We performed expression imputation for each autosome using these tissue weights; thus, we identified genes that were conditionally independent. Based on these 49 tissue weights and a total of 27,977 genes, we used a multiple testing correction of 3.65 × 10−8. If a gene was significant and expressed in at least two tissues, we chose the gene expression with the lowest P value.
Fine-mapping
Fine-mapping of causal gene sets was used to fine-map TWAS at genomic risk regions, identifying likely causal genes77. We used the All-PA–leisure + SSOE EUR meta-analysis and the Vig-PA–leisure as input GWAS, and the GTEx v7 weights from PrediXcan78 combined with Metabolic Syndrome in Men Study79, Netherlands Twins Registry80, Young Finns Study81 and CommonMind Consortium82 weights. LD scores were obtained from the 1000 Genomes Phase 3.
We also conducted fine-mapping analyses at the SNP level using Polygenic functionally informed fine-mapping83, using precomputed prior causal probabilities based on a meta-analysis of 15 UKB traits, allowing for the extraction of per-SNP heritabilities (SNPVAR). These were necessary together with the summary statistics and EUR genotypes from 1000 Genomes Project Phase 3 to perform functionally informed fine-mapping with the sum of single effects84,85.
Multitrait analysis of GWAS (MTAG)
This method was employed to facilitate joint analysis of summary statistics from GWAS of different but related traits, such as the MTAG86, to increase power in GWAS analyses. Here we ran two MTAG analyses, both with the EUR All-PA–leisure + SSOE meta-analysis as a single trait. First, we joined this trait with leisure screen time15. Because of their negative genetic correlation, we flipped the allele in the LSC statistics. Second, we joined the EUR All-PA–leisure + SSOE meta-analysis with a measure of PA-liking87.
Enrichment analysis
g:Profiler is a toolset that also includes finding biological categories enriched in a gene list88. We performed it on three gene lists resulting from the FUMA gene-based test—EUR All-PA–leisure+SSOE meta-analysis, cross-ancestry All-PA–leisure+SSOE meta-analysis and Vig-PA–leisure.
Mitochondrial genome association analyses
We used mitochondria genomes of EUR ancestry individuals from MVP to run mitochondria–SNV association analyses and gene-based association analyses with All-PA–leisure and Vig-PA–leisure phenotypes. For the mitochondria–SNV association analyses, we filtered missing call rates for variants >0.1, missing call rates for samples >0.1, and we excluded monomorphic variants. We thus analyzed 141 SNVs from 189,782 individuals for All-PA–leisure and 142 SNVs from 201,018 individuals for Vig-PA–leisure. For the gene-based association analyses, we initially assigned the SNVs to mitochondrial genes using the reference sequence NC_012920.1 (ref. 89); each SNV mapped to a single gene, and then we used the R software SKAT v2.2.4 to perform associations with All-PA–leisure and Vig-PA–leisure phenotypes, using age, sex and the first ten principal components as covariates90. We considered the nominally significant P value (≤0.05).
PheWAS
We used Vanderbilt University Medical Center’s (VUMC) EHR database to conduct a PheWAS as an empirical exploration of PA and its related phenotypes, investigating a broader range of traits compared to those used in the genetic correlation analyses. The VUMC EHR database, also known as the Synthetic Derivative, houses de-identified, longitudinal medical histories of over 3.1 million patients, including their demographic information (age, sex and ethnicity), and clinical information like medications, lab values, procedural and surgical codes and diagnostic codes27. Codes from both the International Classification of Diseases, ninth and tenth editions (ICD-9 and ICD-10), are used by healthcare professionals to code symptoms, procedures and diagnoses in the synthetic derivative. A subset of the patients from the Synthetic Derivative also has their biological data associated with their medical records. This subset is referred to as BioVU and was used for genomic and phenomic analyses91,92,93.
For our analysis, we performed a PheWAS across BioVU records using a polygenic score for PA derived from the EUR All-PA–leisure + SSOE meta-analysis. First, we used PRS-CS94 to calculate a polygenic score for PA for 66,917 BioVU records of EUR participants with genotyped data. Next, using the PA polygenic score as the continuous predictor, we ran a PheWAS95 to explore the phenomic landscape of PA. The first step in PheWAS involves mapping related ICD codes to their phecode using the R PheWAS package96. The complete list of phecodes, along with the corresponding ICD-9 and ICD-10 codes mapped to them, can be found on the PheWAS catalog ( Finally, using the EHR-driven case-control status for each phecode, we estimated the association between the PA polygenic score and the phecodes. We restricted our analysis to 1,254 phecodes that meet the following three criteria: (1) phecodes need over 100 cases or control, (2) phecodes should not be specific to a single sex and (3) phecodes should not be included in the broader mental health phenotypes. We also controlled for the first ten ancestry principal components, median age of the record and reported sex.
Genomic structural equation modeling
Genomic structural equation modeling (genomic-SEM) was used to evaluate the overall genetic architecture of the PA traits included in the EUR GWAS analyses21. Summary statistics for the All-PA–leisure + SSOE meta-analysis, and for GWAS of Vig-PA–leisure, Vig-PA–home, Vig-PA–work and liking of PA were used to perform EFA and CFA. EFA model fit was evaluated by the amount of cumulative variance explained, the strength of SS loadings (≥1), and balance in the proportion of variance explained by each of the individual factors. Traits with factor loadings ≥0.20 in the EFA were allowed to load on the respective factors and evaluated for CFA model fit as determined by conventional fit indices21.
Survival analyses
We ran the Cox proportional-hazards model using the package survival 3.3-1 in R, to establish the impact of vigorous PA for the three contexts (leisure, home and work) on the risk of dying of any cause. Information was collected in the MVP dataset for age, censoring status, and vigorous PA. Age was calculated for censored individuals from the current year minus the year of birth; for dead participants, the year of death was provided. The vigorous PA phenotypes present in MVP were previously described in the Methods ‘MVP datasets’ as follows: we considered Vig-PA–leisure, Vig-PA–home and Vig-PA–work classification. We had complete information for 252,718 individuals (of whom 55,849 were deceased) for Vig-PA–leisure, 253,096 individuals (of whom 55,203 were deceased) for Vig-PA–home and 207,862 individuals (of whom 43,294 were deceased) for Vig-PA–work.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
link